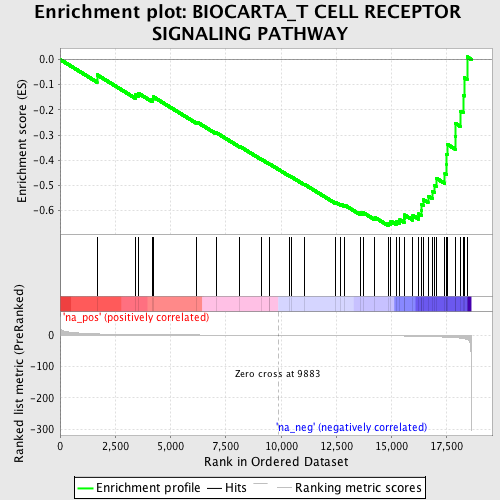
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | BIOCARTA_T CELL RECEPTOR SIGNALING PATHWAY |
| Enrichment Score (ES) | -0.6606882 |
| Normalized Enrichment Score (NES) | -1.7934791 |
| Nominal p-value | 0.013888889 |
| FDR q-value | 0.294262 |
| FWER p-Value | 0.996 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MAPK8 | 1673 | 4.351 | -0.0608 | No | ||
| 2 | ARHGAP5 | 3412 | 2.062 | -0.1405 | No | ||
| 3 | PTPN7 | 3545 | 1.982 | -0.1343 | No | ||
| 4 | SHC1 | 4178 | 1.653 | -0.1572 | No | ||
| 5 | CD4 | 4207 | 1.637 | -0.1477 | No | ||
| 6 | HRAS | 6193 | 0.946 | -0.2482 | No | ||
| 7 | CD3E | 7061 | 0.707 | -0.2901 | No | ||
| 8 | TRB@ | 8136 | 0.423 | -0.3451 | No | ||
| 9 | CD247 | 9117 | 0.190 | -0.3966 | No | ||
| 10 | CAMK2B | 9475 | 0.097 | -0.4151 | No | ||
| 11 | CD3D | 9498 | 0.093 | -0.4157 | No | ||
| 12 | LAT | 10401 | -0.130 | -0.4634 | No | ||
| 13 | FOS | 10490 | -0.151 | -0.4671 | No | ||
| 14 | CD3G | 11065 | -0.311 | -0.4959 | No | ||
| 15 | MAP2K1 | 12474 | -0.704 | -0.5670 | No | ||
| 16 | SOS1 | 12695 | -0.778 | -0.5736 | No | ||
| 17 | PIK3R1 | 12882 | -0.833 | -0.5780 | No | ||
| 18 | PLCG1 | 13591 | -1.102 | -0.6087 | No | ||
| 19 | RAC1 | 13719 | -1.142 | -0.6079 | No | ||
| 20 | RALBP1 | 14250 | -1.393 | -0.6270 | No | ||
| 21 | ZAP70 | 14876 | -1.733 | -0.6490 | Yes | ||
| 22 | PPP3CA | 14970 | -1.793 | -0.6420 | Yes | ||
| 23 | LCK | 15211 | -1.955 | -0.6417 | Yes | ||
| 24 | FYN | 15378 | -2.088 | -0.6366 | Yes | ||
| 25 | PIK3CA | 15572 | -2.276 | -0.6317 | Yes | ||
| 26 | ARHGAP1 | 15585 | -2.289 | -0.6169 | Yes | ||
| 27 | ARHGAP6 | 15966 | -2.700 | -0.6192 | Yes | ||
| 28 | RAF1 | 16233 | -3.067 | -0.6129 | Yes | ||
| 29 | PPP3CB | 16355 | -3.264 | -0.5974 | Yes | ||
| 30 | PPP3CC | 16365 | -3.277 | -0.5759 | Yes | ||
| 31 | MAP2K4 | 16445 | -3.446 | -0.5569 | Yes | ||
| 32 | ARFGAP1 | 16655 | -3.864 | -0.5422 | Yes | ||
| 33 | JUN | 16845 | -4.286 | -0.5235 | Yes | ||
| 34 | PTPRC | 16945 | -4.513 | -0.4985 | Yes | ||
| 35 | ARFGAP3 | 17033 | -4.668 | -0.4717 | Yes | ||
| 36 | RELA | 17416 | -5.813 | -0.4532 | Yes | ||
| 37 | GRB2 | 17489 | -6.003 | -0.4166 | Yes | ||
| 38 | MAPK3 | 17495 | -6.022 | -0.3764 | Yes | ||
| 39 | MAP3K1 | 17523 | -6.133 | -0.3366 | Yes | ||
| 40 | NFKBIA | 17884 | -7.579 | -0.3049 | Yes | ||
| 41 | ARHGAP4 | 17902 | -7.667 | -0.2542 | Yes | ||
| 42 | PRKCA | 18108 | -9.028 | -0.2045 | Yes | ||
| 43 | NCF2 | 18244 | -10.357 | -0.1421 | Yes | ||
| 44 | VAV1 | 18300 | -10.774 | -0.0725 | Yes | ||
| 45 | NFATC1 | 18423 | -13.291 | 0.0104 | Yes |
